Crystalline Organic Molecules and Polymers
In the group of Prof. David Martin at the University of Michigan, Ann Arbor (USA), we have been implementing low-dose HRTEM techniques at high acceleration voltage to directly image the crystalline structure of organic molecules and polymers. Examples illustrating the possibilities of low-dose HRTEM imaging are defects in polyDCHD nanocrystals or multi-scale helical structures in MPDI nanofibers.
1. Direct Imaging of Defects in polyDCHD nanocrystals
Poly[1,6-di(N -carbazolyl)2,4-hexadiyne] (polyDCHD) nanocrystals can be prepared by precipitation from a DCHD solution adding a non-solvent followed by UV irradiation to initiate the polymerization. The resulting polyDCHD nanoparticles are highly crystalline with an average size of about 40 nm x 80 nm.
The polyDCHD nanocrystals are sensitive to the electron beam and the end-point dose for reflections with lattice spacings between 0.3-0.5 nm was measured to be 0.55±0.15 C cm-2 at 400kV. Therefore, low-dose HRTEM imaging conditions on a JEOL 4000EX were choosen to keep the total dose at 0.05-0.1 C cm-2 to prevent noticeable structural changes during data acquisition. An example for the structure of such a nanocrystal with about 25 nm diameter can be seen in Figure 1.1. This crystal exhibits a twin boundary (between red and green) as well as a small angle grain boundary (to the blue part). The different structural representations of the polyDCDHD unit cell in the different parts of the crystal is due to thickness differences between the different parts of the crystal.
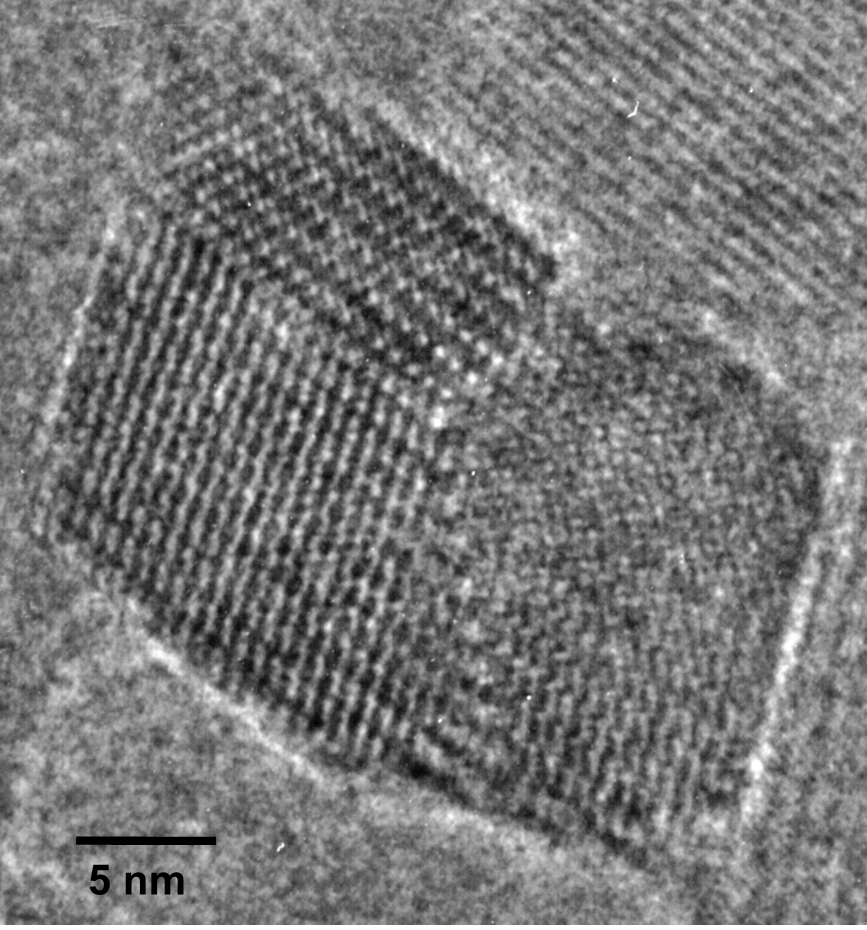
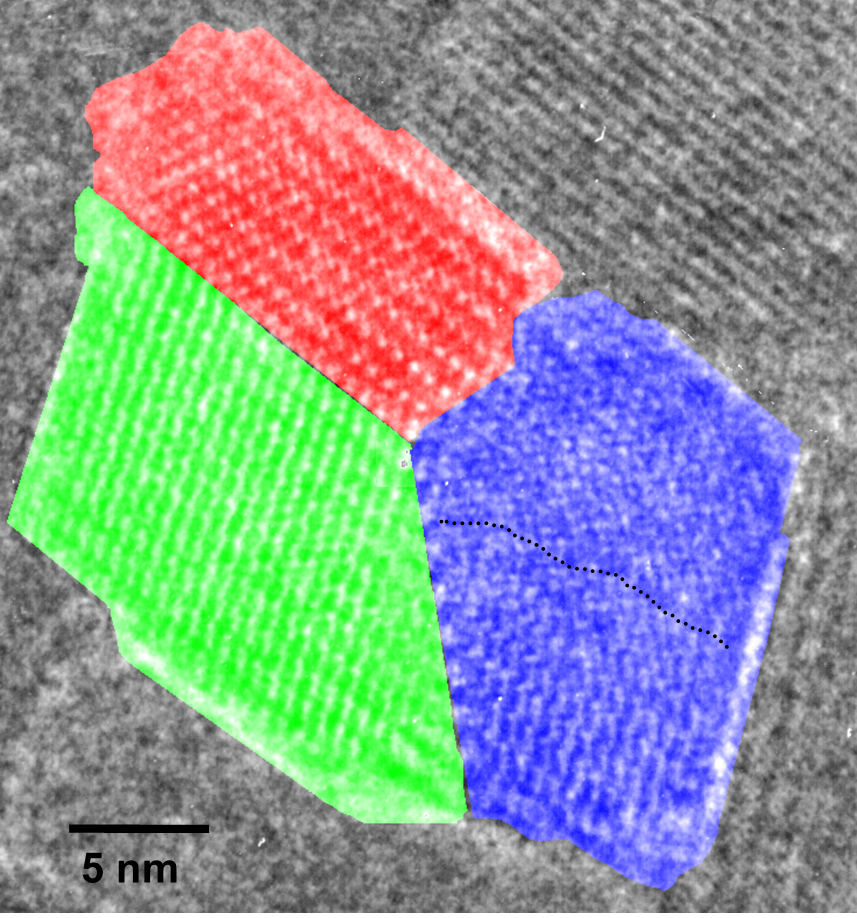
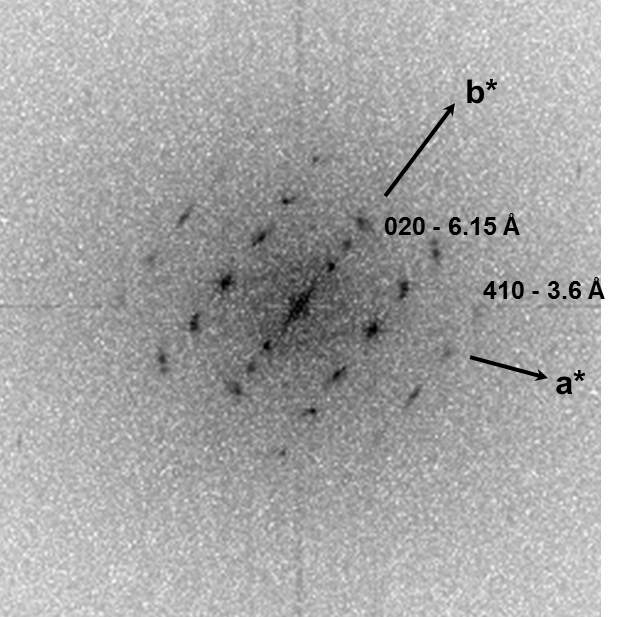
Fig. 1.1: Low-dose HRTEM image of a polyDCHD crystal exhibiting a twin- and a low-angle grain boundary with the different domains indicated by the different colors.
Details of this work have been published at
- C. Kübel, D. C. Martin, "Influence of Structural Variations on High Resolution Electron Microscopy (HREM) Images of Poly[1,6-di(N-carbazolyl)-2,4-hexadiyne] (polyDCHD) Nanocrystals", Phil. Mag. A, 2001, 81, 1651-1673; DOI: 10.1080/01418610010009388.
2. Characterization of the multi-helical structure of MPDI nanofibers
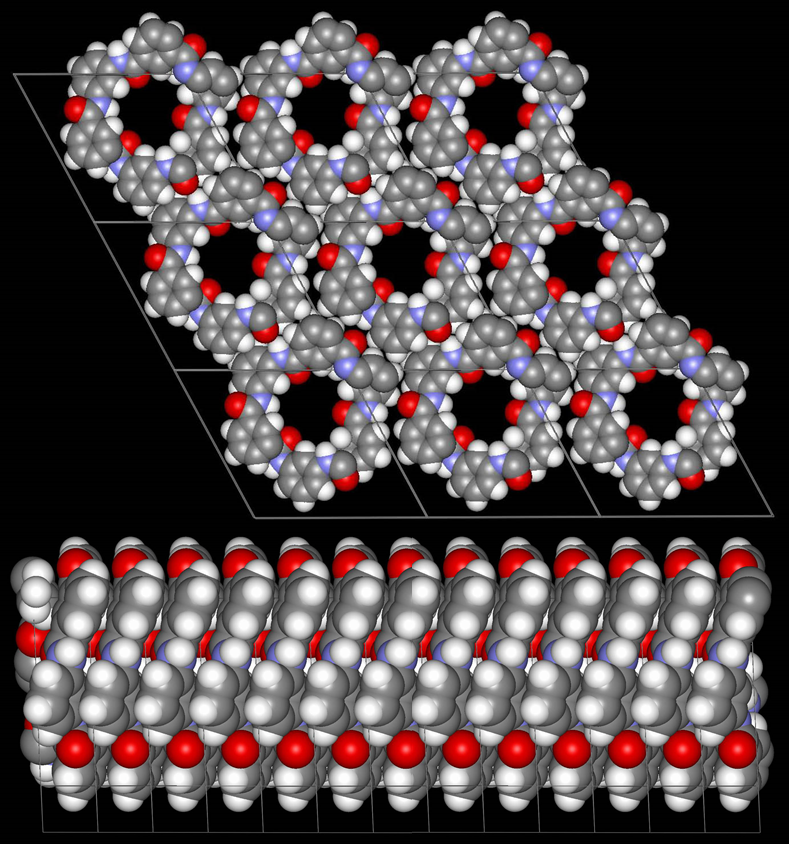
Low-dose-high-resolution electron microscopy (LD-HRTEM) was used to analyze the structure of regularly twisted poly(m-phenylene isophthalamide) (MPDI) strands formed by slow crystallization from solution. The MPDI chains were found to aggregate into regular assemblies exhibiting uniform twisting at several different length scales ranging from the molecular to the mesoscopic. The MPDI polymer backbone formed a flattened helical structure that was organized into twisted bundles. The helical molecular structure promoted good lateral packing, but led to an open core running down the helical axis, which is presumably filled with solvent and calcium chloride. The electron diffraction and
HRTEM data were consistent with a pseudohexagonal unit cell with a = b = 1.65±0.02 nm, gama = 120±0.5°, and c = 0.378 nm, where the c-axis was oriented nominally parallel to the twist axis of the fiber. The strain induced by the mesoscopic twisting is compensated for by lateral shift disorder between the polymer helices, as confirmed by direct HRTEM imaging.

Fig. 2.1. BF-TEM overview image and diffraction pattern of the MPDI nanofibers.
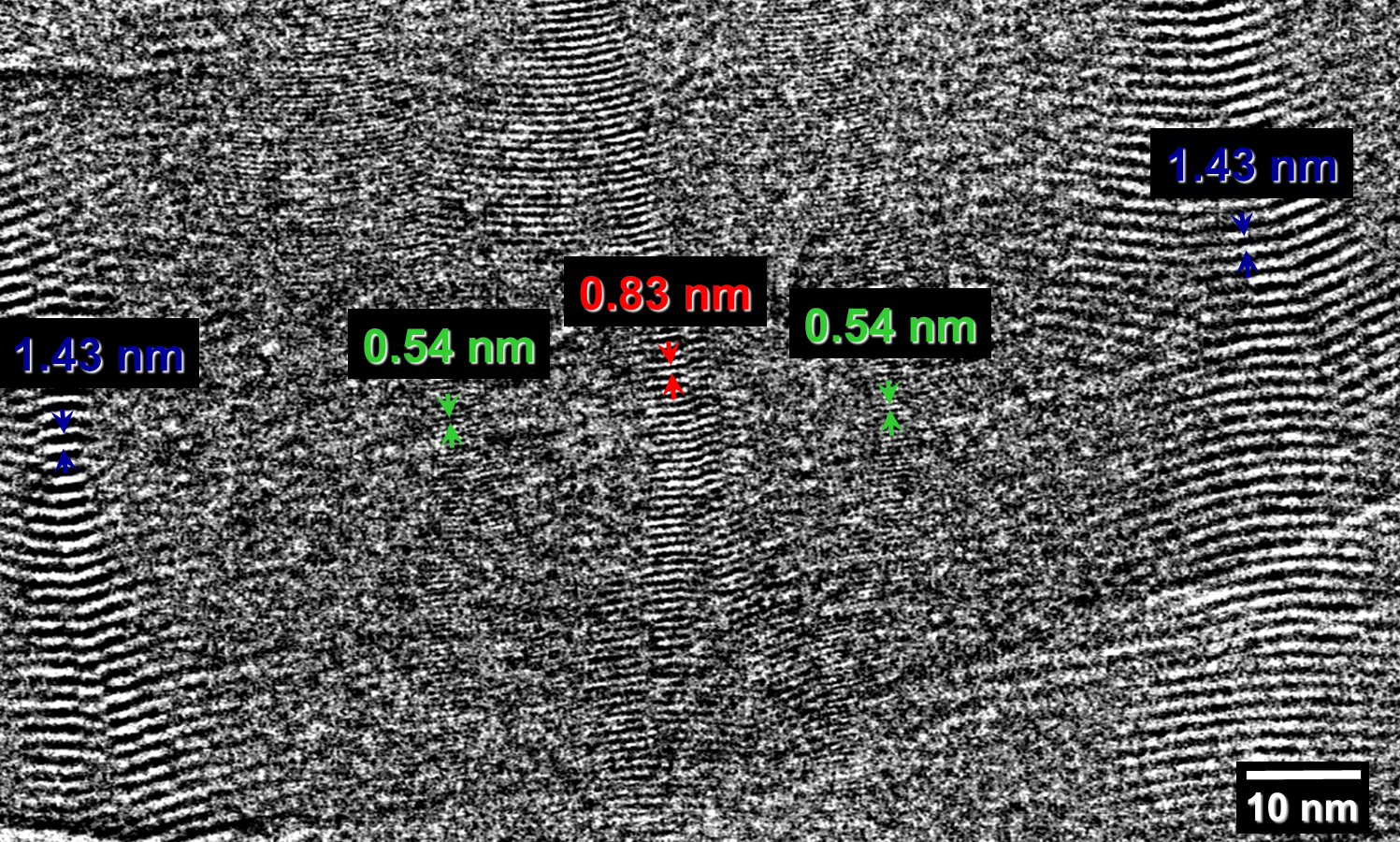
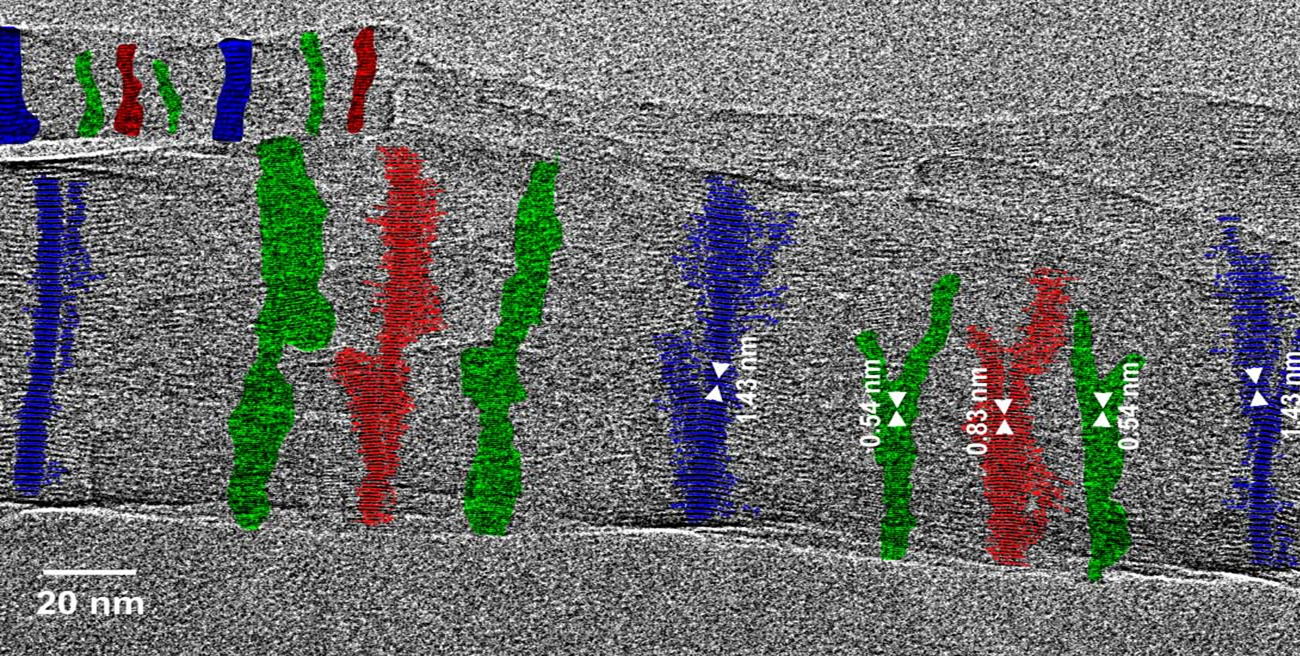
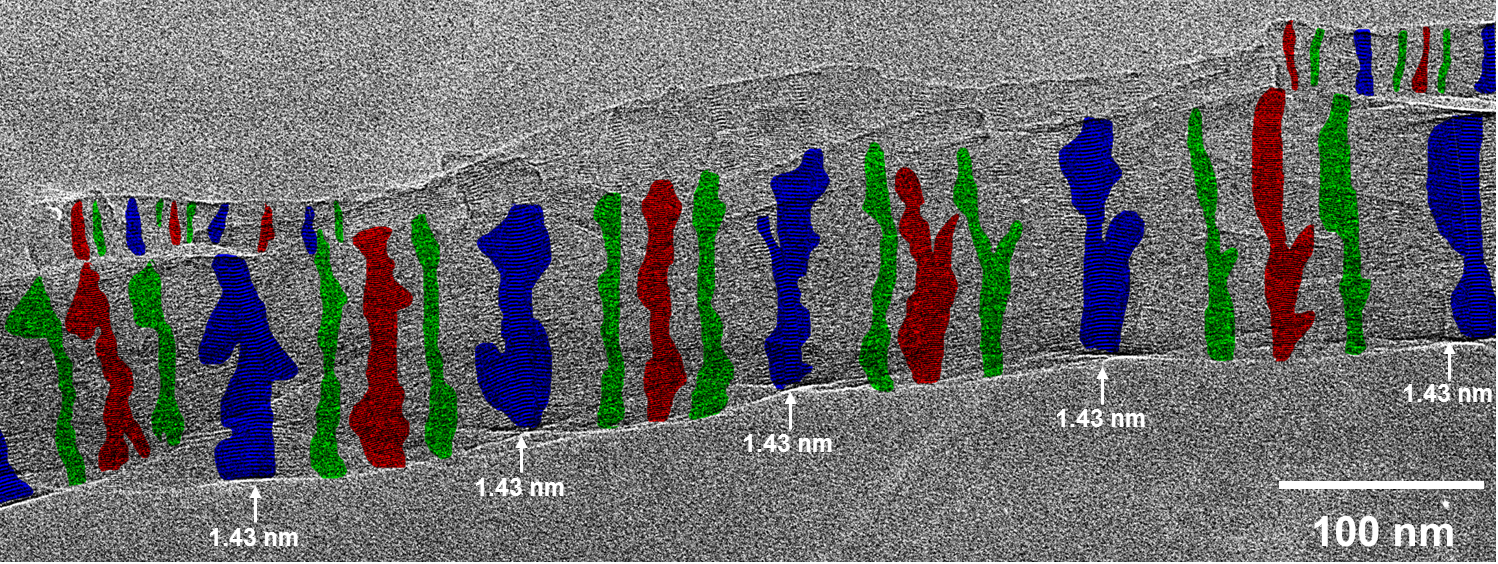
Fig. 2.2: Large-area low-dose HRTEM images of a twisted MPDI nanofiber showing the periodically visible sets of lattice fringes due to the continuous rotation of the crystal orientation along the fiber axis.
|
|
Fig. 2.3: Molecular model in good agreement with the experimentally observed unit cell and schematic representation of the twitsing of the individual MPDI fibrils leading to the observed regular sets of lattice fringes and the mesoscale packing of the individual MPI fibrils into boundles.
Details of this work have been published at
- C. Kübel, D. P. Lawrence, D. C. Martin, "Super-Helically Twisted MPDI Fibers", Macromolecules, 2001, 34(26), 9053-9058.
This work has been performed at the University of Michigan, Ann Arbor with funding from the Alexander von Humboldt foundation and the National Science Foundation.